
Alzheimer's Clinical Research Data via R Packages: the Alzverse
Michael C. Donohue, Kedir Hussen, Oliver Langford, Richard Armenta, Gustavo Jimenez-Maggiora
7 August 2025
Source:vignettes/alzverse-paper.Rmd
alzverse-paper.RmdAbstract
Sharing clinical research data is essential for advancing research in
Alzheimer’s disease (AD) and other therapeutic areas. However,
challenges in data accessibility, standardization, documentation,
usability, and reproducibility continue to impede this goal. In this
article, we highlight the advantages of using R packages to
overcome these challenges using two examples. The first example
R package, ‘A4LEARN’ includes data from a randomized trial
(the Anti-Amyloid Treatment in Asymptomatic Alzheimer’s [A4] study) and
its companion observational study of biomarker negative individuals (the
Longitudinal Evaluation of Amyloid Risk and Neurodegeneration [LEARN]
study). The second example is the ADNIMERGE2 R
package, which includes data from the Alzheimer’s Disease Neuroimaging
Initiative (ADNI). These packages bundle raw and processed data,
documentation, and reproducible analyses into a portable, analysis-ready
formats. By promoting collaboration, transparency, and reproducibility,
R data packages can play a vital role in accelerating
clinical research.
Introduction
Alzheimer’s disease (AD) is one of the leading neurodegenerative diseases worldwide, with growing evidence supporting its multifactorial etiology. The Alzheimer’s Disease Neuroimaging Initiative (ADNI) (Weiner et al. 2025) and similar projects have accumulated vast quantities of clinical, neuroimaging, and biomarker data, creating opportunities for scientific advances in the understanding and treatment of AD. ADNI has provided data for more than 6000 scientific papers publications (Weiner et al. 2025).
Availability of such data is on the rise, due in part to data sharing mandates from funders like the National Institutes of Health. However, it often takes considerable time and effort for researchers to gain sufficient familiarity with the data to produce meaningful analyses. Learning curves can be steep due to inadequate or hard-to-locate documentation and example analysis code.
Open-source software solutions, particularly in the form of
R packages (R Core Team 2019;
Wickham and Bryan 2023), offer significant potential to address
these barriers. The R programming language is widely used
in biostatistics, machine learning, and clinical research.
R packages are a well-known means for distributing cutting
edge statistical software and documentation, and they often include data
and analysis vignettes which demonstrate how the methods can be applied
to data. But R packages can also be used to share data
itself. The authors have maintained the ADNIMERGE R data
package since 2017 ((ADNI) 2023).
ADNIMERGE has been cited by about 250 articles1 and has inspired
related projects, such as the ANMERGE package of AddNeuroMed Consortium
data (Birkenbihl et al. 2021). Vuorre and Crump (2021) demonstrated the utility
of R packages for sharing data and analysis code from
psychological experiments.
We discuss how R packages can also facilitate easy
access, harmonization, and analysis of larger clinical datasets from AD
studies. These packages are built with the goal of providing an audit
trail of derived data progeny, supporting reproducible research, and
leveraging outstanding R tools for unit testing and
validation (Wickham 2011), websites (Wickham, Hesselberth, et al. 2024), and
regulatory submissions (Knoph and contributors
2023).
In this paper we discuss the advantages of using R
packages to share large clinical study datasets, and provide two new
examples: the A4LEARN packages which includes data from a
randomized trial (the Anti-Amyloid Treatment in Asymptomatic Alzheimer’s
[A4] study) (Sperling et al. 2023) and its
companion observational study of biomarker negative individuals (the
Longitudinal Evaluation of Amyloid Risk and Neurodegeneration [LEARN]
study) (Sperling et al. 2024); and the
ADNIMERGE2 package, which includes latest data from the
Alzheimer’s Disease Neuroimaging Initiative (ADNI) (Weiner et al. 2025).
Advantages of R Data Packages
Reproducibility, Portability and Documentation
The most important advantage of using R data packages
for sharing clinical research data, is that it facilitates reproducible
research. R is widely available and free to download (R Core Team 2025), commonly used in statistics
courses, and has active development communities such as academic
statisticians, pharmaceutical statisticians, and commercial enterprises
such as Posit’s RStudio Integrated Development Environment (IDE).
All R packages include a manual that details the
functions and datasets in a standardized format. This content is linked
R object’s name accessible to the R user
(e.g. by typing ?t.test, or ?cars) and can be
browsed within the IDE. This is a drastic improvement compared to the
typical case in which relevant data documentation might be contained in
unlinked documents and/or data dictionary spreadsheets. R
packages also typically provide analysis “vignettes”, which demonstrate
how functions can be applied to available datasets to produce analysis
results as tables or figures. These are also linked and can be browsed
within the IDE. Our A4LEARN package contains a vignette to
exactly reproduce key findings of the published manuscript for the trial
(Sperling et al. 2023). This code can be
used by outside researchers to jump start their own inquiries, and help
ensure the data is being used correctly, efficiently, and consistent
with the intentions of the study team.
The R package bundle of data, R functions,
documentation and vignettes is made portable as an efficiently
compressed file which can be installed on any machine running
R. Data files within the package are also efficiently
compressed using R’s .RData file format. These
.RData files can be read by SPSS, Stata, and SAS [CONFIRM].
Another advantage to .RData compared to tabular text files
(e.g. .csv files), is that they can utilize R
object classes such as dates and factor variables, eliminating the need
to process and annotate data prior to analysis.
The pkgdown R package (Wickham, Hesselberth, et al. 2024) makes it
trivial to export documentation and vignettes as a website, and
integrates well with code repositories such as GitHub. These
pkgdown websites are searchable, and make the documentation
and examples available to researchers who do not use R. See
atri-biostats.github.io/A4LEARN/
and atri-biostats.github.io/ADNIMERGE2
for examples.
Standardized and Efficient Workflow and Testing
Questions often arise about the original source of data or how
derived variables were defined. Therefore, it is crucial to preserve a
record of data progeny. The standardize R package structure
and build workflow makes it easy to retrace the steps of the package
build. R packages also have a standardize framework for
testing (Wickham 2011) and tools for
“assertive” programming to verify assumptions about the data (Fischetti 2023).
The R package structure and workflow has been well-documented (Wickham and Bryan 2023). We briefly review the structure while highlighting some key aspects in the context of the clinical research data.
data-raw. The data-raw
directory is intended to house raw data and code to import and process
raw data and store as .Rdata files in the data
directory. Raw data can be preserved in the package with minimal
manipulation, or it can be processed attaching meta data (variable
labels and units), and ensuring factors and dates are stored as the
correct object class. Data dictionary spreadsheets can be parsed to
provide content for manual pages (Wickham,
Danenberg, et al. 2024).
vignettes. The vignettes
directory houses the analysis demonstrations, typically as Rmarkdown
(.Rmd) files (Allaire et al.
2024). We prefer to create derived datasets and variables as a
vignette, as well, so that derivations are easily accessible to
researchers within the IDE. These vignettes can include assertive
programming to ensure data conforms to expectations (Fischetti 2023). ADNIMERGE2
contains vignettes which use pharmaverse workflows to derive
CDISC ADaM datasets.
R. The R directory
contains .R files with code defining R
functions and manual content (Wickham, Danenberg,
et al. 2024). This directory can store scoring functions, which
might be necessary to derive scores from item-level data from
psychometric assessments, for example.
testthat. The testthat
directory includes automated tests that are checked when the package is
built. Sensitive and crucial code that requires replication by
independent programmers can be tested here, to ensure they produce
equivalent results on the actual data and/or test data.
reports. Report code that is not wanted
as a vignette can be stored in a separate directory for general reports.
Of note, rmarkdown supports “parameterized reports”, which
can produce different output depending on the supplied parameter(s).
Clinical trials like A4 often include several outcomes collected on the
same schedule and analyzed with the same approach. Instead of writing
identical code for several outcomes, one generic parameterized rmarkdown
file can produce all of these reports. Clinical trial outcomes are often
aggregated into one long dataset with a row for each subject, time
point, and outcome (see ADQS in the A4LEARN
package). The parameterized report can filter this long dataset for the
desired outcome and analyze only that outcome. Futhermore, using
R parallel programming tools (e.g. Wickham (2023)), these reports can be produced
in parallel. In the case of the A4 trial read out, once data from the
blinded phase was locked and unblinded it only took about 30 minutes to
build the final data package and render all planned analysis reports and
summary slide decks.
Example R Data Packages
ADNIMERGE2
The ADNIMERGE2 package was built using pharmaverse tools. It can be downloaded from loni.usc.edu. Below are examples of some basic summaries of participant characteristics by phase, or wave, of ADNI can be created using the derived ADSL data table in the package.
tbl_summary(
data = ADNIMERGE2::ADSL %>%
filter(ENRLFL %in% "Y"),
by = ORIGPROT,
include = c(AGE, SEX, EDUC, RACE, ETHNIC, BMI, DX, APOE,
ADASTT13, CDRSB, MMSCORE),
type = all_continuous() ~ "continuous2",
statistic = list(
all_continuous() ~ "{mean} ({sd})",
all_categorical() ~ "{n} ({p}%)"
),
digits = all_continuous() ~ 1,
percent = "column",
missing_text = "(Missing)"
) %>%
add_overall(last = TRUE) %>%
add_stat_label(label = all_continuous2() ~ "Mean (SD)") %>%
modify_footnote_header(
footnote = "Column-wise percentage; n (%)",
columns = all_stat_cols(),
replace = TRUE
) %>%
modify_abbreviation(abbreviation = "ADNI: Alzheimer’s Disease Neuroimaging Initiative; CN: Cognitive Normal; MCI: Mild Cognitive Impairment; DEM: Dementia.") %>%
modify_caption(
caption = "Table 1. ADNI - Subject Characteristics by Study Phase"
) %>%
bold_labels()| Characteristic |
ADNI1 N = 8191 |
ADNIGO N = 1311 |
ADNI2 N = 7901 |
ADNI3 N = 6961 |
ADNI4 N = 4771 |
Overall N = 2,9131 |
|---|---|---|---|---|---|---|
| Age (in Years) | ||||||
| Mean (SD) | 75.2 (6.8) | 71.6 (7.9) | 72.7 (7.2) | 70.7 (7.4) | 68.7 (7.4) | 72.2 (7.6) |
| Sex, n (%) | ||||||
| Female | 342 (42%) | 60 (46%) | 379 (48%) | 381 (55%) | 307 (64%) | 1,469 (50%) |
| Male | 477 (58%) | 71 (54%) | 411 (52%) | 315 (45%) | 170 (36%) | 1,444 (50%) |
| Education | ||||||
| Mean (SD) | 15.5 (3.0) | 15.8 (2.7) | 16.3 (2.6) | 16.4 (2.3) | 16.0 (2.8) | 16.0 (2.7) |
| (Missing) | 1 | 0 | 0 | 0 | 1 | 2 |
| Race, n (%) | ||||||
| American Indian or Alaskan Native | 1 (0.1%) | 1 (0.8%) | 1 (0.1%) | 2 (0.3%) | 3 (0.6%) | 8 (0.3%) |
| Asian | 14 (1.7%) | 1 (0.8%) | 14 (1.8%) | 29 (4.2%) | 45 (9.4%) | 103 (3.5%) |
| Black or African American | 39 (4.8%) | 4 (3.1%) | 34 (4.3%) | 105 (15%) | 158 (33%) | 340 (12%) |
| Native Hawaiian or Other Pacific Islander | 0 (0%) | 0 (0%) | 2 (0.3%) | 1 (0.1%) | 0 (0%) | 3 (0.1%) |
| Other Pacific Islander | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 2 (0.4%) | 2 (<0.1%) |
| White | 762 (93%) | 118 (90%) | 728 (92%) | 537 (77%) | 237 (50%) | 2,382 (82%) |
| More than one race | 3 (0.4%) | 5 (3.8%) | 10 (1.3%) | 13 (1.9%) | 22 (4.6%) | 53 (1.8%) |
| Unknown | 0 (0%) | 2 (1.5%) | 1 (0.1%) | 9 (1.3%) | 10 (2.1%) | 22 (0.8%) |
| Ethnicity, n (%) | ||||||
| Hispanic or Latino | 19 (2.3%) | 8 (6.1%) | 31 (3.9%) | 58 (8.3%) | 66 (14%) | 182 (6.2%) |
| Not Hispanic or Latino | 794 (97%) | 122 (93%) | 755 (96%) | 637 (92%) | 409 (86%) | 2,717 (93%) |
| Unknown | 6 (0.7%) | 1 (0.8%) | 4 (0.5%) | 1 (0.1%) | 2 (0.4%) | 14 (0.5%) |
| Body Mass Index | ||||||
| Mean (SD) | 27.6 (4.1) | 24.0 (NA) | 30.8 (7.2) | 30.0 (NA) | 29.3 (6.5) | 29.2 (6.2) |
| (Missing) | 815 | 130 | 784 | 695 | 450 | 2,874 |
| Baseline Diagnostics Status, n (%) | ||||||
| CN | 225 (29%) | 1 (1.0%) | 295 (37%) | 380 (55%) | 292 (61%) | 1,193 (42%) |
| MCI | 373 (48%) | 99 (99%) | 344 (44%) | 243 (35%) | 148 (31%) | 1,207 (42%) |
| DEM | 182 (23%) | 0 (0%) | 151 (19%) | 73 (10%) | 37 (7.8%) | 443 (16%) |
| (Missing) | 39 | 31 | 0 | 0 | 0 | 70 |
| APOE Genotype, n (%) | ||||||
| ε2/ε2 | 2 (0.2%) | 0 (0%) | 3 (0.4%) | 1 (0.1%) | 2 (1.7%) | 8 (0.3%) |
| ε2/ε3 | 53 (6.5%) | 9 (7.0%) | 66 (8.5%) | 52 (7.7%) | 6 (5.1%) | 186 (7.4%) |
| ε2/ε4 | 18 (2.2%) | 2 (1.6%) | 14 (1.8%) | 17 (2.5%) | 4 (3.4%) | 55 (2.2%) |
| ε3/ε3 | 363 (44%) | 67 (52%) | 352 (45%) | 347 (52%) | 53 (45%) | 1,182 (47%) |
| ε3/ε4 | 295 (36%) | 42 (33%) | 269 (35%) | 204 (30%) | 46 (39%) | 856 (34%) |
| ε4/ε4 | 88 (11%) | 8 (6.3%) | 75 (9.6%) | 52 (7.7%) | 7 (5.9%) | 230 (9.1%) |
| (Missing) | 0 | 3 | 11 | 23 | 359 | 396 |
| Baseline ADAS-Cog Item 13 Total Score | ||||||
| Mean (SD) | 18.4 (9.2) | 12.4 (5.4) | 16.1 (10.1) | 13.1 (8.9) | 13.0 (8.1) | 15.4 (9.4) |
| (Missing) | 8 | 1 | 7 | 11 | 39 | 66 |
| Baseline CDR Sum of Boxes Score | ||||||
| Mean (SD) | 1.8 (1.8) | 1.2 (0.7) | 1.5 (1.9) | 1.0 (1.6) | 0.9 (1.5) | 1.4 (1.7) |
| (Missing) | 2 | 0 | 0 | 0 | 0 | 2 |
| Baseline MMSE Score | ||||||
| Mean (SD) | 26.7 (2.7) | 28.3 (1.5) | 27.4 (2.7) | 28.0 (2.5) | 27.9 (2.4) | 27.5 (2.6) |
| (Missing) | 2 | 0 | 0 | 0 | 3 | 5 |
| Abbreviation: ADNI: Alzheimer’s Disease Neuroimaging Initiative; CN: Cognitive Normal; MCI: Mild Cognitive Impairment; DEM: Dementia. | ||||||
| 1 Column-wise percentage; n (%) | ||||||
# Prepare analysis dataset of ADAS-cog item-13 score
ADADAS <- ADNIMERGE2::ADQS %>%
# Enrolled participant
filter(ENRLFL %in% "Y") %>%
# ADAS-cog item-13 total score
filter(PARAMCD %in% "ADASTT13") %>%
mutate(TIME = convert_number_days(ADY, unit = 'year')) %>%
filter(!if_any(all_of(c("TIME", "DX", "AVAL")), ~ is.na(.x)))
# Individual profile (spaghetti) plot
ggplot(ADADAS, aes(x = TIME, y = AVAL, group = USUBJID, color = DX)) +
geom_line(alpha = 0.5) +
scale_color_manual(values = c("#73C186", "#F2B974", "#DF957C", "#999999")) +
labs(
y = "ADAS-cog13 Total Score",
x = "Years since baseline visit",
color = "Baseline Diagnostics Status") +
theme(legend.position = "bottom") +
guides(colour = guide_legend(override.aes = list(alpha = 1)))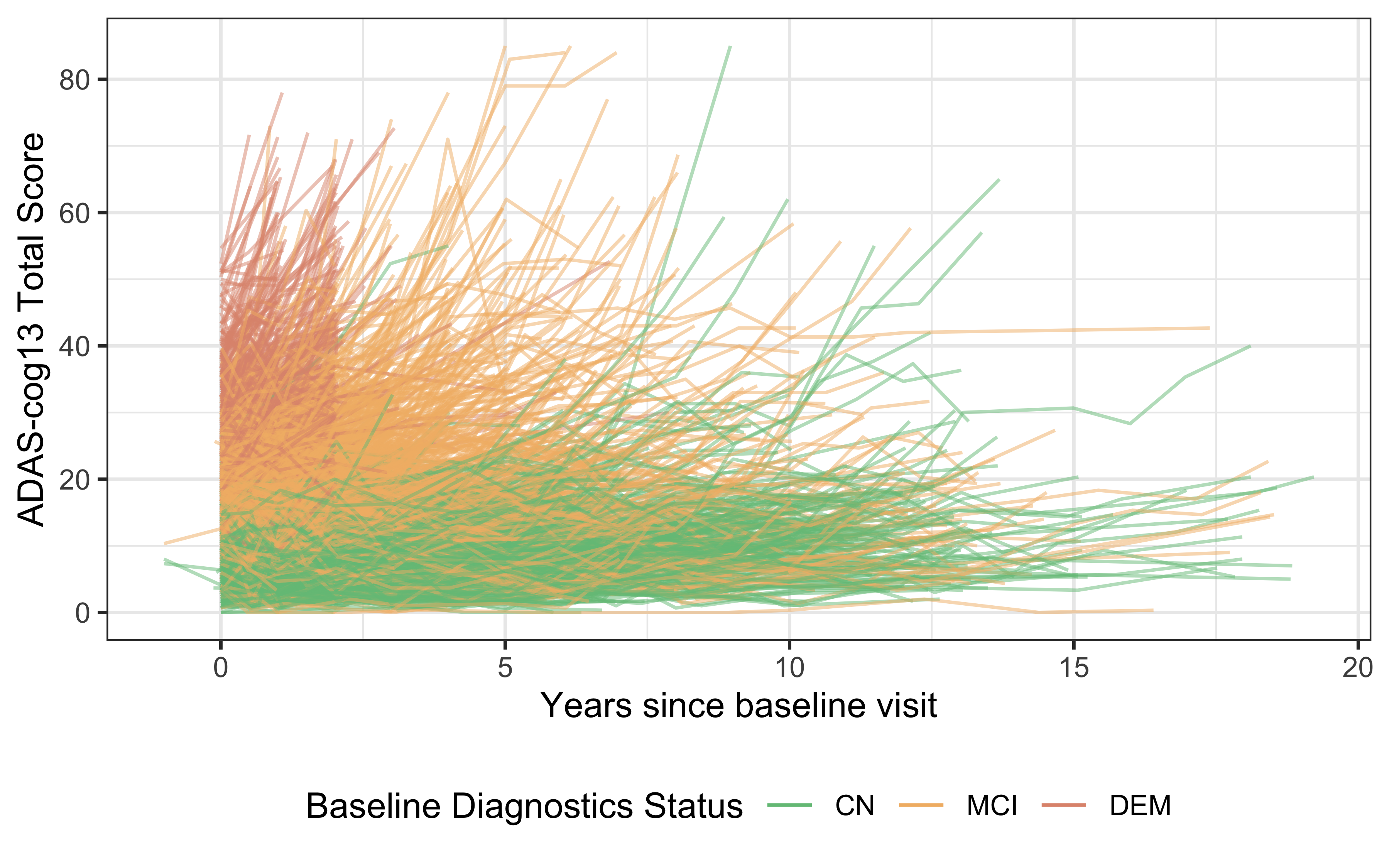
Figure 1. Spahetti plot of ADAS-cog13 scores in ADNI by baseline clinical diagnosis.
A4LEARN
Similarly, A4LEARN data can be easily summarized
tbl_summary(
data = A4LEARN::SUBJINFO %>% filter(SUBSTUDY != 'SF'),
by = SUBSTUDY,
label = SUBJINFO_labels,
include = c(AGEYR, SEX, EDCCNTU, RACE, ETHNIC, BMIBL, SUBSTUDY, APOEGN,
PACCV6, MMSETSV6, AMYLCENT),
type = all_continuous() ~ "continuous2",
statistic = list(
all_continuous() ~ "{mean} ({sd})",
all_categorical() ~ "{n} ({p}%)"
),
digits = all_continuous() ~ 1,
percent = "column",
missing_text = "(Missing)"
) %>%
add_overall(last = TRUE) %>%
add_stat_label(label = all_continuous2() ~ "Mean (SD)") %>%
modify_footnote_header(
footnote = "Column-wise percentage; n (%)",
columns = all_stat_cols(),
replace = TRUE
) %>%
modify_abbreviation(abbreviation = "A4: Anti-Amyloid Treatment in Asymptomatic Alzheimer’s study; LEARN: Longitudinal Evaluation of Amyloid Risk and Neurodegeneration.") %>%
modify_caption(
caption = "Table 2. A4 and LEARN - Subject Characteristics by substudy."
) %>%
bold_labels()| Characteristic |
A4 N = 1,1691 |
LEARN N = 5381 |
Overall N = 1,7071 |
|---|---|---|---|
| Age in years at Consent | |||
| Mean (SD) | 71.9 (4.8) | 70.5 (4.3) | 71.5 (4.7) |
| Sex, n (%) | |||
| Male | 475 (41%) | 208 (39%) | 683 (40%) |
| Female | 694 (59%) | 330 (61%) | 1,024 (60%) |
| Years of education | |||
| Mean (SD) | 16.6 (2.8) | 16.8 (2.6) | 16.6 (2.8) |
| Race, n (%) | |||
| American Indian or Alaskan Native | 2 (0.2%) | 5 (0.9%) | 7 (0.4%) |
| Asian | 24 (2.1%) | 12 (2.2%) | 36 (2.1%) |
| Black or African American | 28 (2.4%) | 14 (2.6%) | 42 (2.5%) |
| More than one race | 8 (0.7%) | 5 (0.9%) | 13 (0.8%) |
| Unknown or Not Reported | 7 (0.6%) | 1 (0.2%) | 8 (0.5%) |
| White | 1,100 (94%) | 501 (93%) | 1,601 (94%) |
| Ethnicity, n (%) | |||
| Hispanic or Latino | 34 (2.9%) | 18 (3.3%) | 52 (3.0%) |
| Not Hispanic or Latino | 1,124 (96%) | 516 (96%) | 1,640 (96%) |
| Unknown or Not reported | 11 (0.9%) | 4 (0.7%) | 15 (0.9%) |
| body mass index (weight (kg) / [height (m)]^2) | |||
| Mean (SD) | 27.4 (5.1) | 27.6 (4.9) | 27.4 (5.0) |
| (Missing) | 2 | 1 | 3 |
| APOE4 genotype (ε2/ε4, ε3/ε4, ε4/ε4, no ε4), n (%) | |||
| E2/E2 | 2 (0.2%) | 5 (0.9%) | 7 (0.4%) |
| E2/E3 | 61 (5.2%) | 66 (12%) | 127 (7.4%) |
| E2/E4 | 35 (3.0%) | 10 (1.9%) | 45 (2.6%) |
| E3/E3 | 417 (36%) | 342 (64%) | 759 (45%) |
| E3/E4 | 560 (48%) | 111 (21%) | 671 (39%) |
| E4/E4 | 94 (8.0%) | 2 (0.4%) | 96 (5.6%) |
| (Missing) | 0 | 2 | 2 |
| PACC Total Score at Visit 6 | |||
| Mean (SD) | 0.0 (2.7) | NA (NA) | 0.0 (2.7) |
| (Missing) | 0 | 538 | 538 |
| MMSE Total Score at Visit 6, n (%) | |||
| 22 | 1 (<0.1%) | 0 (NA%) | 1 (<0.1%) |
| 24 | 5 (0.4%) | 0 (NA%) | 5 (0.4%) |
| 25 | 25 (2.1%) | 0 (NA%) | 25 (2.1%) |
| 26 | 36 (3.1%) | 0 (NA%) | 36 (3.1%) |
| 27 | 103 (8.8%) | 0 (NA%) | 103 (8.8%) |
| 28 | 229 (20%) | 0 (NA%) | 229 (20%) |
| 29 | 347 (30%) | 0 (NA%) | 347 (30%) |
| 30 | 419 (36%) | 0 (NA%) | 419 (36%) |
| (Missing) | 4 | 538 | 542 |
| Amyloid PET centiloids. AMYLCENT = 183.07 x SUVRCER – 177.26. | |||
| Mean (SD) | 66.1 (32.8) | 4.2 (12.6) | 46.6 (40.2) |
| Abbreviation: A4: Anti-Amyloid Treatment in Asymptomatic Alzheimer’s study; LEARN: Longitudinal Evaluation of Amyloid Risk and Neurodegeneration. | |||
| 1 Column-wise percentage; n (%) | |||
Merging A4, LEARN, and ADNI
ADQS_meta <- ADNIMERGE2::ADQS %>%
filter(ENRLFL %in% "Y") %>%
bind_rows(A4LEARN::ADQS %>%
select(STUDYID = SUBSTUDY, USUBJID = BID, PARAMCD = QSTESTCD,
AVAL = QSSTRESN, ADY = QSDTC_DAYS_T0)) %>%
mutate(TIME = convert_number_days(ADY, unit = 'year'))
ADQS_meta %>%
filter(DX == 'CN' | STUDYID %in% c('A4', 'LEARN'),
PARAMCD %in% c('MMSCORE', 'MMSE')) %>%
ggplot(aes(x = TIME, y = AVAL, color = STUDYID)) +
geom_line(aes(group = USUBJID), alpha = 0.25) +
labs(
y = "MMSE Total Score",
x = "Years since baseline visit",
color = "Study") +
theme(legend.position = "bottom") +
guides(colour = guide_legend(override.aes = list(alpha = 1)))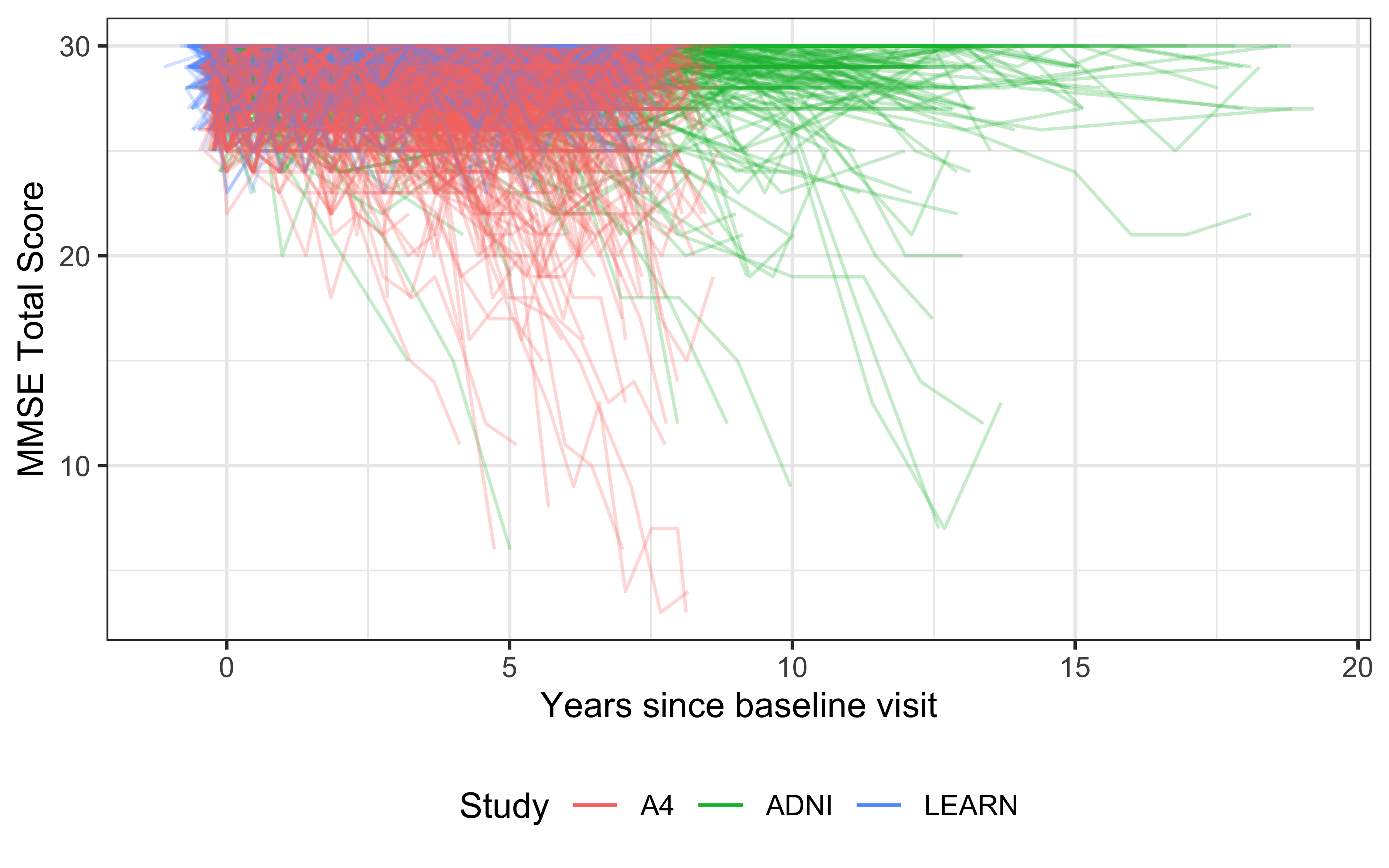
Figure 2. Spahetti plot of MMSE scores in ADNI CN, A4, and LEARN.
Discussion
The development of A4LEARN and ADNIMERGE2
represents a step forward in enabling the Alzheimer’s disease research
community to share and analyze data more effectively, and serve as a
template for additional future study packages. These packages facilitate
the transition from proprietary software like SAS to open-source tools,
allowing greater flexibility and transparency in the research process.
The shift from SAS to R represents a broader trend in the
clinical research community toward open-source and reproducible research
practices.
Challenges remain, particularly in the area of data access. In our examples data packages can be sourced using the existing data access models. This puts the onus on data users to go to different sites to obtain data. Once retrieved, The use of locally installed packages poses potential risks, which can be mitigated by using containerization and package management tools like Docker and renv for version control.
Methods
The primary data sources for the packages are derived from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) the A4 and LEARN companions studies. These datasets include longitudinal clinical and neuroimaging data, cognitive test scores, genetic and biomarker data, and other modalities that have been harmonized to facilitate cross-study comparisons.
To ensure usability and consistency, we curated the datasets by mapping variables to standardized terminologies (e.g., CDISC ADaM, SDTM), handling missing data through imputation techniques, and deriving key analysis variables.
A4LEARN and ADNIMERGE2 were developed using
best practices in R package development, including:
- R Package Architecture: Each package is modular, supporting various stages of data analysis, from raw data processing to the generation of regulatory-compliant datasets.
- Data Standardization: The packages support standardization of clinical data, with built-in functions to harmonize variable formats, handle missing values, and generate standardized metadata.
- Reproducibility: Built-in vignettes and examples guide users through
the installation, data loading, and analysis processes. The packages
integrate with tools like
renvand Docker to facilitate reproducibility in different computing environments.
The R package framework facilitates the creation of
Analysis Data Model (ADaM) datasets, which are the gold standard for
statistical analysis in clinical trials. The admiral package is used to
generate ADaM datasets for ADNIMERGE2. Other pharmaverse tools can be
used to create regulatory-compliant tables, listings, and figures.
Data Availability
Data is available from:
-
A4LEARN: A4StudyData.org -
ADNIMERGE2: loni.usc.edu
Data documentation is available from
-
A4LEARN: atri-biostats.github.io/A4LEARN/ -
ADNIMERGE2: atri-biostats.github.io/ADNIMERGE2
Code Availability
R code is available for download from the following repositories:
- alzverse: github.com/atri-biostats/alzverse
- A4LEARN: github.com/atri-biostats/A4LEARN
- ADNIMERGE2: github.com/atri-biostats/ADNIMERGE2